ISCB 2018 Poster
Metalloproteome Landscape From the Amino Acid Covariance Perspective
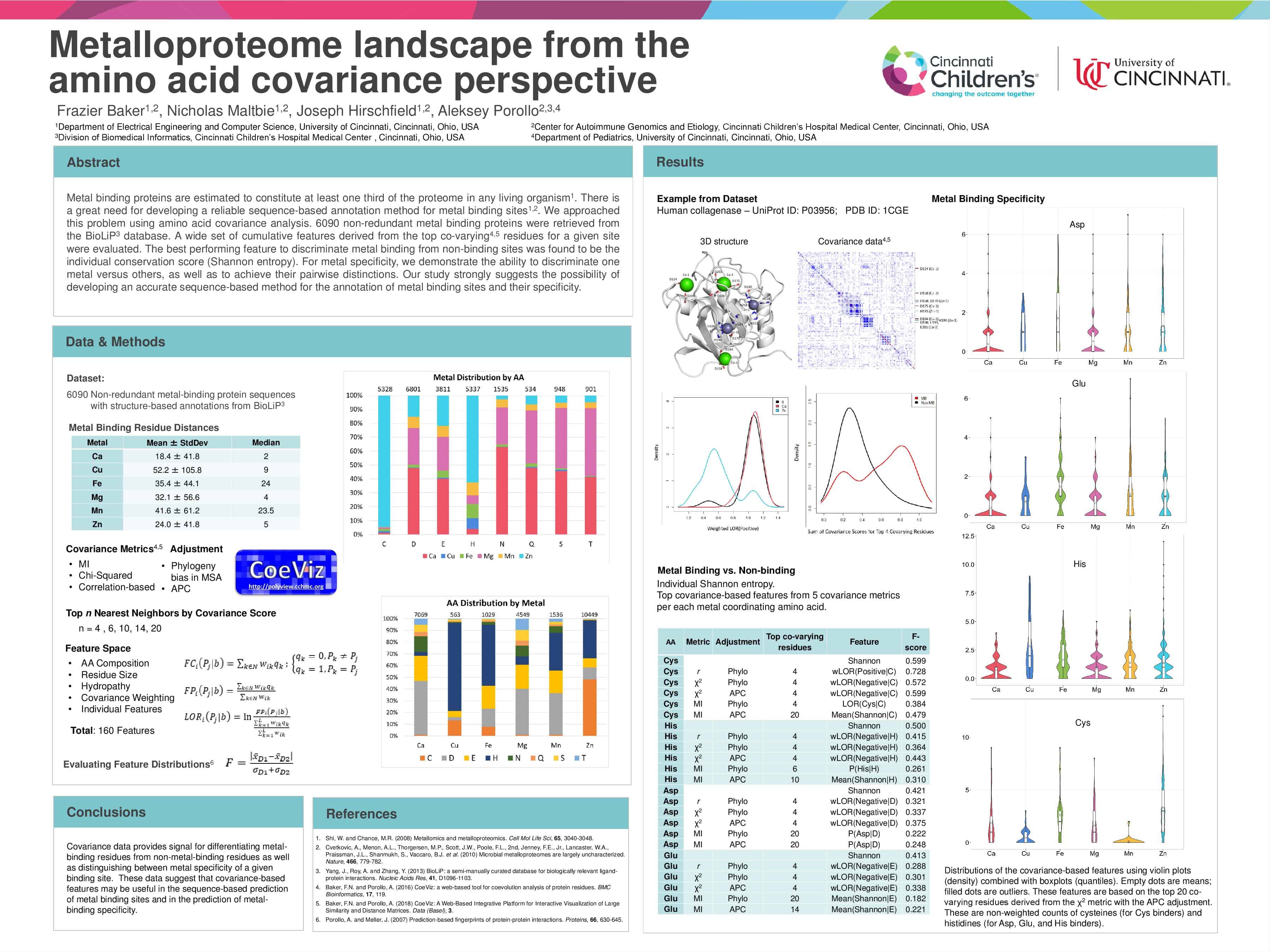
Poster presented at 2018 Intelligent Systems for Molecular Biology Conference. The poster is listed on the website’s poster list.
Short Abstract
Metal binding proteins are estimated to constitute at least one third of the proteome in any living organism. There is a great need for developing a reliable sequence-based annotation method for metal binding sites. We approached this problem using amino acid covariance analysis. 6090 non-redundant metal binding proteins were retrieved from the BioLiP database. A wide set of cumulative features derived from the top co-varying residues for a given site were evaluated. The best performing feature to discriminate metal binding from non-binding sites was found to be the individual conservation score (Shannon entropy). For metal specificity, the correlation-based metric appears the most informative to discriminate one metal versus others, as well as to achieve their pairwise distinctions. When discerning one type of metal from the other five types, metals can be discriminated in the following descending order of signal strength: Zn > Cu > Ca > Mg > Mn > Fe. In pairwise comparisons, Ca vs Mg appears to be the hardest metal pair to discern. Our study strongly suggests the possibility of developing an accurate sequence-based method for the annotation of metal binding sites and their specificity.